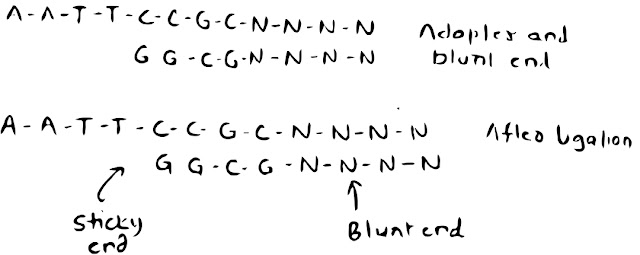Recombinant DNA Technology (RDT): The technique that enable the isolation, identification and recombination of DNA from different sources so that new unique characteristics can be introduced into an organism.
RDT is also known as genetic engineering.
The enzymes used in recombinant DNA technology fall into four broad categories.
Template dependent DNA polymerase
DNA polymerase enzyme that synthesises new polynuclear dice complimentary to the existing DNA or RNA templates are included in this category.
Different types of DNA polymerase are used in gene manipulation.
- DNA polymerase I (Kornberg Enzyme): First DNA polymerase enzyme discovered in E. coli by Arthur Kornberg.
It is DNA dependent DNA polymerase.
It Possesses three enzymatic activities.
By splitting the enzyme DNA polymerase with protease (Subtilisin or Trypsin) gives,
1) Small N-terminal fragment: 5’-3’ polymerase activity——Gap filling and DNA repairing.
2) Large C-terminal fragment: 3’-5’ and 5’-3’ exonuclease activities (Klenow fragment) ——Proofreading activity, Primar removal.
- Reverse Transcriptase (RTase): Synthesize DNA from RNA template.
It is RNA dependent DNA polymerase.
Discovered by Howard Temin and David Baltimore independently at a same time.
- Taq DNA polymerase: A DNA polymerase isolated from a thermostable bacterium, Thermus aquaticus.
stable at high temperature at 90°C.
Activities:
5'-3' polymerase activity
5'-3' exonuclease proofreading activity.
3'-5' exonuclease activity (Lack off)
Nucleases
Nuclease enzyme degrades nucleic acids by breaking the phosphodiester bonds that holds the nucleotides together.
Nucleases are two different kinds.
- Exonuclease: That remove nucleotides at the end (Either 3'- or 5'-) of nucleic acid by breaking phosphodiester bond.
- Endonuclease: That break internal phosphodiester bond (in between 3'- to 5'-) within a nucleic acid.
Some examples of Nucleases.
- Mug bean Nuclease: Endonuclease, isolated from Mung Bean sprouts.
specific to ssDNA and RNA, leave dsDNA intact.
catalytic activity: Zn+2
- S1 nuclease: Endonuclease, isolated from Aspergillus Oryzae.
Degrade ssDNA or RNA, not degrade dsDNA and RNA-DNA hybrid.
Cleave complementary strand opposite to nick.
digest ssRNA at 3'- end of pyrimidine residues.
digest only the RNA strand of an RNA-DNA heteroduplex.
- Restriction endonuclease:
Restriction enzymes are defined as the enzyme which cut DNA at defined sites (recognition or restriction site).
Present in Bacterial cells, as a part of Protective mechanism called "Restriction Mechanism" system. In this system, the restriction enzymes hydrolyse any exogenous DNA (Viral attack) that is introduced into cell.
It does not act on host cell's DNA because Methylase (a modification enzyme) modifies particular bases in the recognition sequence and prevents the restriction enzyme from cutting the DNA.
Restriction enzymes classified into Three types,
Type I, Type II and Type III.
| Characteristics |
Type I |
Type II |
Type III |
| Nature of Enzyme |
Both endonuclease and methylase activity |
Uni functional enzyme: Endonuclease; Methylase activity |
Bifunctional: Endonuclease and Methylase activity |
| Restriction Requires |
Mg²⁺ + ATP + S-adenosyl methionine |
Mg²⁺ |
Mg²⁺ |
| Cleavage Site |
Random |
At or near recognition site |
≈ 25 bp from recognition site |
| Example |
EcoB |
EcoRI |
EcoP1 |
Most of the enzymes used today are type II.
The existence of Restriction Enzyme was first postulated by W. Arber. he noticed when DNA of bacteriophage inter into host bacterium in cut up into smaller pieces.
In 1970, Hamilton Smith and Co- worker first isolated a restriction enzyme from bacterium Haemephilus influenzae, Hind II (six cutter).
Nomenclature: Three Parts
- Abbreviation of genus and species to three letters. e.g. E from genus Escherichia and co from species Coli.
- A letter, number or combination of two to indicate Strain.
- A Roman numeral to indicate the order in which different Restriction modification system was found.
Example: EcoRI
E Escherichia (Genus)
co Coli (Species)
R RY13 (Strain)
I First identified
Restriction Site: A definite length of dsDNA segment that contains particular nucleotide sequence of 4, 6 or 8 base pair length from restriction enzyme cuts DNA, known as Restriction or Recognition site.
These are generally Palindromic Sequences.
Symbol of enzyme cut = "/"
Restriction enzymes cut DNA strand into Two ways.
- Blunt end: cleave both strands at same point of axis of symmetry.
- Overhanging end: Cleave points on both strand of DNA in different symmetry. (also called Sticky or cohesive end)
staggered cut gives either 5'- overhanging end or 3'- overhanging end.
NB: The restriction site for EcoRI enzyme is 5'-GAATTC-3'.
Frequency of Recognition Points: The Number of Recognition site for particular Restriction endonuclease in a DNA molecule of known length can be measured mathematically.
The expected frequency of particular sequences = 1/4^n
Uses of Restriction Endonuclease:
- A proper amount of enzyme is added to intended DNA in a buffer solution and the reaction is heated at 27°C.
Restriction Mapping: The technique which is used to get the information about recognition site for different enzymes on DNA sample is known as Restriction Mapping. It involves cutting of DNA with R.E then formed fragments are checked for their size on Agarose Gel.
On the basis of cleaveage done by restriction enzyme, Restriction enzymes are,
- Isoschizomers: A pair of R.E that recognize the same recognition sequences and cut in the same location.
Examples: SphI (CGTAC/G) and BbuI (CGTAC/G)
- Neoschizomers: Recognise same sequence, cut it differently.
Examples: SmaI (GGG/CCC) and XmaI (G/GGCCC)
- Isocaudomers: Recognise slightly different sequence but produce the same ends.
Examples: Sau3A (N/GATC) and BamHI (G/GATCC)
End modifications Enzymes:
End modification in Enzyme make changes to the ends of DNA molecules.
- Terminal deoxynucleotidyl transferase: Template independent DNA polymerase, able to synthesise a new DNA nucleotide without base pairing of incoming nucleotides to end existing strand of DNA or RNA.
This enzyme is used for formation of a cohesive end by Homopolymer tailing (addition of same nucleotides) at 3’-OH termini of a dsDNA.
- Alkaline phosphatase: Remove phosphate groups from 5’ ends of DNA.
Obtained from bacteria (E. coli) and calf intestinal tissue.
- T4 polynucleotide kinase: Perform reverse reaction to alkaline phosphatase.
Adding phosphates to 5’ ends of DNA (end labelling).
Ligase:
DNA ligases join DNA molecule together by synthesising Phosphodiester bond between nucleotides at the end of two different molecules or two end of a single molecule.
Obtained from E. coli or bacteriophage T4.
Linkers and Adaptors:
Linker: linkers are short stretches of dsDNA of Known nucleotide sequence(8-14bps) And having recognition site 3-8 restriction enzymes.
Linkers are ligated to blunt ends of DNA by ligase. After ligation, cohesive ends are generated by digesting the DNA with proper restriction enzymes.
Problem with linkers: Sites for the enzymes used to generate cohesive ends may be present in the target DNA fragments.
Adaptor: Adaptors are linkers with cohesive ends.
Blunt end of DNA with blunt end of Adaptors joined by ligase, gives sticky ends.